nf-core/proteinfold
Protein 3D structure prediction pipeline
Introduction
This document describes the user-facing output produced by the pipeline.
Pipeline overview
The pipeline is built using Nextflow and predicts protein structures using the following methods:
See main README.md for a condensed overview of the steps in the pipeline, and the bioinformatics tools used at each step.
The directories listed below will be created in the output directory after the pipeline has finished. All paths are relative to the top-level results directory.
Exact subdirectories depend on the selected mode(s). In a multi-mode run (for example alphafold2,boltz,rosettafold_all_atom) you will typically see top-level directories such as alphafold2/, boltz/, rosettafold_all_atom/, multiqc/, reports/, compare/, and pipeline_info/.
Prediction outputs (all modes)
User-facing outputs are largely consistent across modes.
Common output patterns
<MODE>/top_ranked_structures/<SEQUENCE NAME>.pdb<MODE>/<SEQUENCE NAME>/<SEQUENCE NAME>_plddt.tsv<MODE>/<SEQUENCE NAME>/paes/<SEQUENCE NAME>_<RANK>_pae.tsv(when available)<MODE>/<SEQUENCE NAME>/<SEQUENCE NAME>_*msa.tsv(mode-specific MSA summary)<MODE>/<SEQUENCE NAME>/<SEQUENCE NAME>_{ptm,iptm}.tsvand chainwise summaries (where applicable)
Canonical .tsv metric formats (including pLDDT, MSA, (i)pTM, chain-wise (i)pTM and PAE) are defined in the contributor documentation: Processable structure prediction metrics.
Example report plots
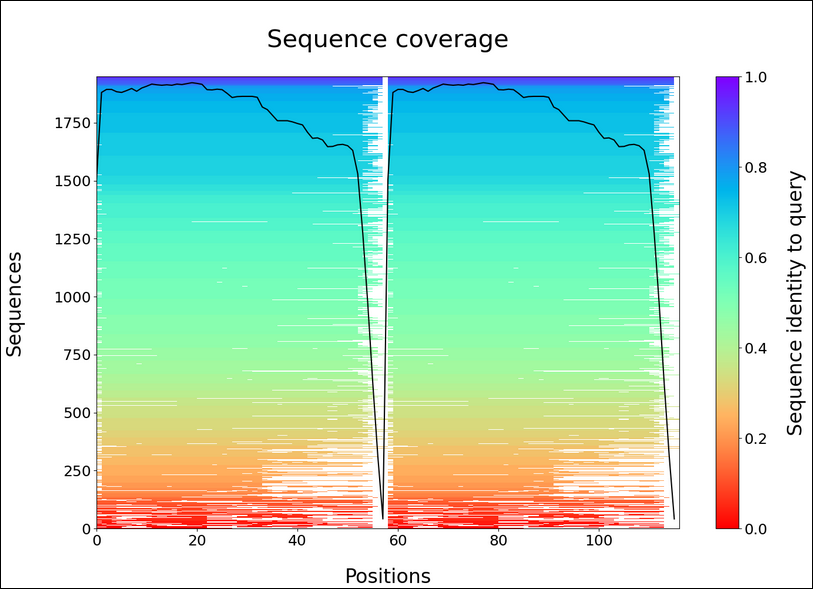
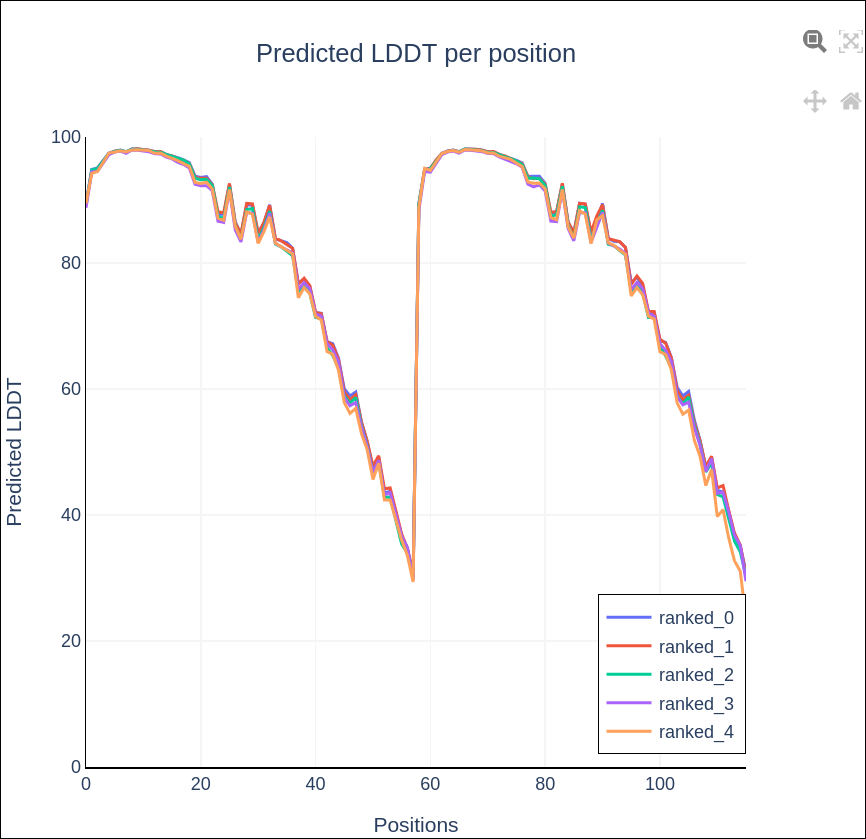
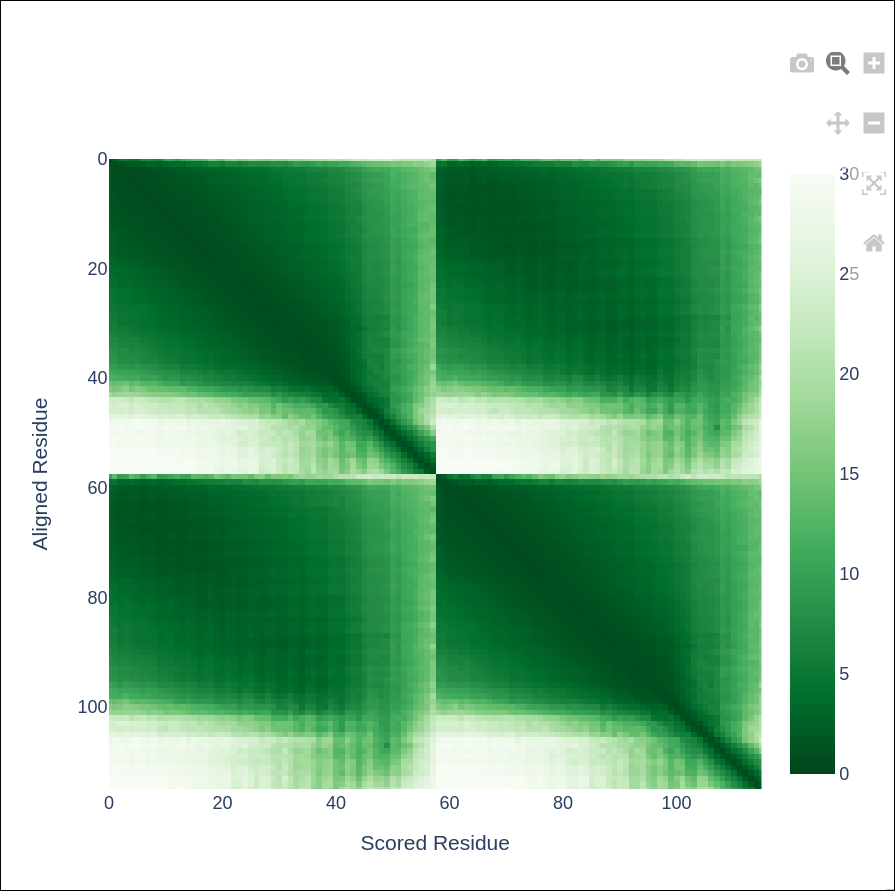
The report exports include key visualisations such as sequence coverage, predicted Local Distance Difference Test (pLDDT), and Predicted Aligned Error (PAE).
Sequence coverage
predicted Local Distance Difference Test (pLDDT)
Predicted Aligned Error (PAE)
Per-mode reports and comparisons
Output files
reports/<SEQUENCE NAME>_<MODE>_report.html(single-mode report per sequence/mode)
compare/<SEQUENCE NAME>_comparison_report.html(present when running multiple modes)
MultiQC report
Output files
multiqc*_multiqc_report.html: Standalone HTML report(s) that can be viewed in your web browser.*_multiqc_report_data/: Parsed report data for each corresponding MultiQC report.
MultiQC is a visualisation tool that generates HTML report(s) summarising samples in your project. Most QC results are visualised in the report and further statistics are available within each corresponding *_multiqc_report_data/ directory.
Results generated by MultiQC collate QC metrics from the selected structure-prediction mode(s), and the software versions for traceability. For more information about how to use MultiQC reports, see http://multiqc.info.
Pipeline information
Output files
pipeline_info/- Reports generated by Nextflow:
execution_report.html,execution_timeline.html,execution_trace.txtandpipeline_dag.dot/pipeline_dag.svg. - Reports generated by the pipeline:
pipeline_report.html,pipeline_report.txtandsoftware_versions.yml. Thepipeline_report*files will only be present if the--email/--email_on_failparameter’s are used when running the pipeline. - Reformatted samplesheet files used as input to the pipeline:
samplesheet.valid.csv. - Parameters used by the pipeline run:
params.json.
- Reports generated by Nextflow:
Nextflow provides excellent functionality for generating various reports relevant to the running and execution of the pipeline. This will allow you to troubleshoot errors with the running of the pipeline, and also provide you with other information such as launch commands, run times and resource usage.
Additional intermediate outputs
Depending on the selected mode(s) and options, additional top-level directories may be present, for example:
fasta2yaml/(for YAML conversion inputs/outputs)mmseqs/results/(for MMseqs2 outputs such as.a3mfiles)split/output_msa/(for split-MSA intermediate CSV outputs)
--save_intermediates
If --save_intermediates is enabled, extra raw intermediate files are published in mode-specific raw/ directories.
Examples include:
alphafold2/<MODE>/<SEQUENCE NAME>/raw/colabfold/<SEQUENCE NAME>/raw/boltz/<SEQUENCE NAME>/boltz_results_*/rosettafold_all_atom/<SEQUENCE NAME>/raw/alphafold3/<SEQUENCE NAME>/raw/helixfold3/<SEQUENCE NAME>/raw/rosettafold2na/<SEQUENCE NAME>/raw/
These raw outputs are intended for advanced debugging, reproducibility and method-specific downstream analyses. For detailed, canonical tool-specific native output specifications, see: